Difference between revisions of "General Information/The casposases"
Jump to navigation
Jump to search
| (3 intermediate revisions by the same user not shown) | |
(No difference)
| |
Latest revision as of 12:51, 5 December 2022
The casposases
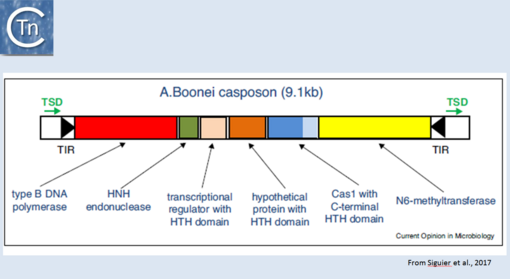
More recently, TE related to CRISPRs, Casposons have been identified[1] and a reassement of their ends has led to the identification of an 14-15 bp target duplication [2]. Moreover, the purified Cas1 enzyme encoded by these ancestral transposons has been demonstrated to catalyse strand transfer of a pre-cleaved transposon in vitro but does not appear to promote transposon strand cleavage in this assay[3]. Cas1 "casposases" use similar chemistry to that used by the CRISPR Cas1-Cas2 complex but with opposite substrate specificities since CRISPR Cas1-Cas2 integrates "random" sequences into a specific site in the CRISPR locus whereas casposases integrate a specific site (the casposon ends) into random target sequences (Fig.24.1).

Fig.24.1. Organization of the Aciduliprofundum boonei casposon. The casposon is shown as an unfilled box. Black filled arrow-heads represent the terminal inverted repeats (TIR); green arrows, the target site duplication (TSD); the genes are as follows: type B DNA polymerase (red box); HNH endonuclease (green); transcriptional regulator with HTH domain (pink); hypothetical protein with HTH domain (orange); cas1 (dark blue) with a c-terminal HTH (light blue); N6-methyltransferase (yellow). From Hickman and Dyda (2015). Non-CRISPR-associated cas1 form two distinct clades. One contains three distinct casposon families (1, 2, and 3) which share features with eukaryotic Polinton/Maverick eukaryotic transposons labeled ‘self-synthesizing’ since they include B family DNA polymerase genes. Members of the three families differ in gene content and evolutionary provenance of the DNA polymerases (protein-primed or RNA-primed).
Bibliography
- ↑ Krupovic M, Makarova KS, Forterre P, Prangishvili D, Koonin EV . Casposons: a new superfamily of self-synthesizing DNA transposons at the origin of prokaryotic CRISPR-Cas immunity. - BMC Biol: 2014 May 19, 12;36 [PubMed:24884953] [DOI]
- ↑ Hickman AB, Dyda F . CRISPR-Cas immunity and mobile DNA: a new superfamily of DNA transposons encoding a Cas1 endonuclease. - Mob DNA: 2014, 5;23 [PubMed:25180049] [DOI]
- ↑