Difference between revisions of "General Information/Non-autonomous IS derivatives"
| Line 1: | Line 1: | ||
{| | {| | ||
| − | |'''<big>M</big>'''any prokaryotic genomes are littered with IS fragments and small, non-autonomous IS derivatives whose transposition can, in principle, be catalyzed in trans by the Tpase of a related complete IS [[:Image:1.13.1.png|(Fig. | + | |'''<big>M</big>'''any prokaryotic genomes are littered with IS fragments and small, non-autonomous IS derivatives whose transposition can, in principle, be catalyzed in trans by the Tpase of a related complete IS [[:Image:1.13.1.png|(Fig.8.1)]]. Structures called MITEs ('''M'''iniature '''I'''nverted repeat '''T'''ransposable '''E'''lements), first identified in ''Neisseria''<ref name=":0"><pubmed>2842323</pubmed></nowiki></ref> are found in many organisms including plants<ref><nowiki><pubmed>12618411</pubmed></nowiki></ref><ref name=":1"><pubmed>11756687</pubmed></nowiki></ref><ref><nowiki><pubmed>1332797</pubmed></nowiki></ref><ref><nowiki><pubmed>8061524</pubmed></nowiki></ref>, insects<ref><nowiki><pubmed>16507338</pubmed></nowiki></ref><ref><nowiki><pubmed>16919158</pubmed></nowiki></ref>, fungi<ref><nowiki><pubmed>17179071</pubmed></nowiki></ref> bacteria <ref name=":0" /><ref><nowiki><pubmed>19393167</pubmed></nowiki></ref><ref><nowiki><pubmed>22016774</pubmed></nowiki></ref><ref><nowiki><pubmed>31085513</pubmed></nowiki></ref><ref><nowiki><pubmed>26442174</pubmed></nowiki></ref> and archaea<ref><nowiki><pubmed>15046567</pubmed></nowiki></ref><ref name=":2"><pubmed>11814653</pubmed></nowiki></ref><ref><nowiki><pubmed>16935554</pubmed></nowiki></ref> are related to IS with DDE Tpases<ref name=":2" /><ref name=":3"><pubmed>17347521</pubmed></nowiki></ref>. Among those derived from IS with DDE transposases, representatives of the [[IS Families/IS1 family|IS''1'']], [[IS Families/IS4 and related families|IS''4'']]'','' [[IS Families/IS5 and related IS1182 families|IS''5'']], [[IS Families/IS6 family|IS''6'']], and even [[Transposons families/Tn3 family|Tn''3'' family]] members such as IS''Rf1'' from ''[[wikipedia:Sinorhizobium_fredii|Sinorhizobium fredii]]'' have been identified. They are generally less than 300 bp long, include appropriately oriented left and right terminal IR but no Tpase and generally generate flanking DR. MITEs are probably derived from IS by internal deletion. Some carry short non-coding sequences between these IRs which may or may not be IS-derived. MITEs are considered to be non-autonomous transposable elements mobilizable in trans by Tpases of full-length parental genomic copies. They were first identified in plants<ref name=":1" /><ref><nowiki><pubmed>15020481</pubmed></nowiki></ref><ref><nowiki><pubmed>15831788</pubmed></nowiki></ref><ref><nowiki><pubmed>17578919</pubmed></nowiki></ref> and are related to Tc/mariner elements (distantly related to bacterial [[IS Families/IS630 family|IS''630'' family]]). IS''630''-related MITEs were also the first described bacterial examples<ref name=":0" /><ref><nowiki><pubmed>11707339</pubmed></nowiki></ref><ref><nowiki><pubmed>12095618</pubmed></nowiki></ref><ref><nowiki><pubmed>21283790</pubmed></nowiki></ref><ref><nowiki><pubmed>10537186</pubmed></nowiki></ref>. MITEs showing similarities to other IS and transposon families are observed in bacteria and archaea. Those related to the [[Transposons families/Tn3 family|Tn''3'' family]] have been called TIMEs<ref><nowiki><pubmed>25121765</pubmed></nowiki></ref>. |
| − | MITE-like structures related to elements with other types of transposase have also been identified. Among these are [[IS Families/IS200-IS605 family|IS''200''/IS''605'' family]] derivatives<ref name=":3" />; ([https://scholar.google.fr/citations?user=WHAtfqcAAAAJ&hl=fr P. Siguier], unpublished) [[:Image:1.18.1.png|(Fig. | + | MITE-like structures related to elements with other types of transposase have also been identified. Among these are [[IS Families/IS200-IS605 family|IS''200''/IS''605'' family]] derivatives<ref name=":3" />; ([https://scholar.google.fr/citations?user=WHAtfqcAAAAJ&hl=fr P. Siguier], unpublished) [[:Image:1.18.1.png|(Fig.13.1)]] now called PATEs ('''P'''alindrome-'''A'''ssociated '''T'''ransposable '''E'''lements)<ref><nowiki><pubmed>21701686</pubmed></nowiki></ref> reflecting the sub-terminal secondary structures which constitute the ends of these IS. Although originally observed in the Archaea as derivatives of known [[IS Families/IS200-IS605 family|IS''200''/IS''605'' family]] members, they have also been observed in certain cyanobacteria and ''[[wikipedia:Salmonella|Salmonella]]'' ([https://scholar.google.fr/citations?user=WHAtfqcAAAAJ&hl=fr P. Siguier] unpublished). They presumably represent decay products that appear quite frequently for this IS family. |
Full-length copies of the parental IS may not be available or may be so divergent as to escape detection in standard [https://blast.ncbi.nlm.nih.gov/Blast.cgi BLAST analysi]s. This is the case for certain MITEs from the archaea <ref><nowiki><pubmed>X</pubmed></nowiki></ref> and is probably also true for the bacteria. Their detection and analysis are therefore arduous. | Full-length copies of the parental IS may not be available or may be so divergent as to escape detection in standard [https://blast.ncbi.nlm.nih.gov/Blast.cgi BLAST analysi]s. This is the case for certain MITEs from the archaea <ref><nowiki><pubmed>X</pubmed></nowiki></ref> and is probably also true for the bacteria. Their detection and analysis are therefore arduous. | ||
| − | Another group of IS derivatives related to MITEs are called MICs ('''M'''obile '''I'''nsertion '''C'''assette)<ref name=":4"><pubmed>15228527</pubmed></nowiki></ref><ref><nowiki><pubmed>10320586</pubmed></nowiki></ref> [[:Image:1.13.1.png|(Fig. | + | Another group of IS derivatives related to MITEs are called MICs ('''M'''obile '''I'''nsertion '''C'''assette)<ref name=":4"><pubmed>15228527</pubmed></nowiki></ref><ref><nowiki><pubmed>10320586</pubmed></nowiki></ref> [[:Image:1.13.1.png|(Fig.8.1)]]. These, like MITEs, are flanked by IR, do not include a Tpase gene and generate flanking DR. They carry various coding sequences, and are present in relatively low copy number. The [[IS Families/IS4 and related families#IS231|IS''231'' sub-group]] of the large [[IS Families/IS4 and related families|IS''4'' family]] includes examples of many of these IS-derivatives (canonical ISs, MITEs, MICs and tIS)<ref name=":4" /><ref><nowiki><pubmed>18215304</pubmed></nowiki></ref>. |
| Line 12: | Line 12: | ||
<br /> | <br /> | ||
| − | |[[Image:1.18.1.png|thumb|450x450px|'''Fig | + | |[[Image:1.18.1.png|thumb|450x450px|'''Fig.13.1.''' PATEs ('''P'''alindrome-'''A'''ssociated '''T'''ransposable '''E'''lements) isolated ends from IS''609''. |
The particular case of MITES derived from IS200/IS605 family members. Top: The IS ends with their essential secondary structures are shown in red (left end) and blue (right end). The boxed (left) sequence represents the conserved target sequence and underlined (right) sequence at the right end are sequences recognized by guide sequences at the foot of the secondary structures necessary for directing cleavage. The inset represents a full-sized IS with its transposase tnpA1 (red) and the accessory gene tnpB (pale green). The actual DNA sequences are shown at the bottom of the figure.|alt=]]<br /> | The particular case of MITES derived from IS200/IS605 family members. Top: The IS ends with their essential secondary structures are shown in red (left end) and blue (right end). The boxed (left) sequence represents the conserved target sequence and underlined (right) sequence at the right end are sequences recognized by guide sequences at the foot of the secondary structures necessary for directing cleavage. The inset represents a full-sized IS with its transposase tnpA1 (red) and the accessory gene tnpB (pale green). The actual DNA sequences are shown at the bottom of the figure.|alt=]]<br /> | ||
Revision as of 12:56, 23 June 2020
| Many prokaryotic genomes are littered with IS fragments and small, non-autonomous IS derivatives whose transposition can, in principle, be catalyzed in trans by the Tpase of a related complete IS (Fig.8.1). Structures called MITEs (Miniature Inverted repeat Transposable Elements), first identified in Neisseria[1] are found in many organisms including plants[2][3][4][5], insects[6][7], fungi[8] bacteria [1][9][10][11][12] and archaea[13][14][15] are related to IS with DDE Tpases[14][16]. Among those derived from IS with DDE transposases, representatives of the IS1, IS4, IS5, IS6, and even Tn3 family members such as ISRf1 from Sinorhizobium fredii have been identified. They are generally less than 300 bp long, include appropriately oriented left and right terminal IR but no Tpase and generally generate flanking DR. MITEs are probably derived from IS by internal deletion. Some carry short non-coding sequences between these IRs which may or may not be IS-derived. MITEs are considered to be non-autonomous transposable elements mobilizable in trans by Tpases of full-length parental genomic copies. They were first identified in plants[3][17][18][19] and are related to Tc/mariner elements (distantly related to bacterial IS630 family). IS630-related MITEs were also the first described bacterial examples[1][20][21][22][23]. MITEs showing similarities to other IS and transposon families are observed in bacteria and archaea. Those related to the Tn3 family have been called TIMEs[24].
MITE-like structures related to elements with other types of transposase have also been identified. Among these are IS200/IS605 family derivatives[16]; (P. Siguier, unpublished) (Fig.13.1) now called PATEs (Palindrome-Associated Transposable Elements)[25] reflecting the sub-terminal secondary structures which constitute the ends of these IS. Although originally observed in the Archaea as derivatives of known IS200/IS605 family members, they have also been observed in certain cyanobacteria and Salmonella (P. Siguier unpublished). They presumably represent decay products that appear quite frequently for this IS family. Full-length copies of the parental IS may not be available or may be so divergent as to escape detection in standard BLAST analysis. This is the case for certain MITEs from the archaea [26] and is probably also true for the bacteria. Their detection and analysis are therefore arduous. Another group of IS derivatives related to MITEs are called MICs (Mobile Insertion Cassette)[27][28] (Fig.8.1). These, like MITEs, are flanked by IR, do not include a Tpase gene and generate flanking DR. They carry various coding sequences, and are present in relatively low copy number. The IS231 sub-group of the large IS4 family includes examples of many of these IS-derivatives (canonical ISs, MITEs, MICs and tIS)[27][29].
|
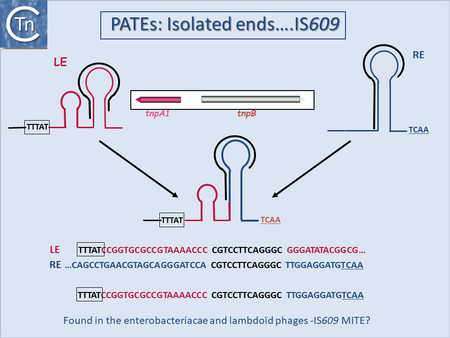 Fig.13.1. PATEs (Palindrome-Associated Transposable Elements) isolated ends from IS609. The particular case of MITES derived from IS200/IS605 family members. Top: The IS ends with their essential secondary structures are shown in red (left end) and blue (right end). The boxed (left) sequence represents the conserved target sequence and underlined (right) sequence at the right end are sequences recognized by guide sequences at the foot of the secondary structures necessary for directing cleavage. The inset represents a full-sized IS with its transposase tnpA1 (red) and the accessory gene tnpB (pale green). The actual DNA sequences are shown at the bottom of the figure. |
Bibliography
- ↑ 1.0 1.1 1.2 </nowiki>
- ↑ <pubmed>12618411</pubmed>
- ↑ 3.0 3.1 </nowiki>
- ↑ <pubmed>1332797</pubmed>
- ↑ <pubmed>8061524</pubmed>
- ↑ <pubmed>16507338</pubmed>
- ↑ <pubmed>16919158</pubmed>
- ↑ <pubmed>17179071</pubmed>
- ↑ <pubmed>19393167</pubmed>
- ↑ <pubmed>22016774</pubmed>
- ↑ <pubmed>31085513</pubmed>
- ↑ <pubmed>26442174</pubmed>
- ↑ <pubmed>15046567</pubmed>
- ↑ 14.0 14.1 Brügger K, Redder P, She Q, Confalonieri F, Zivanovic Y, Garrett RA . Mobile elements in archaeal genomes. - FEMS Microbiol Lett: 2002 Jan 10, 206(2);131-41 [PubMed:11814653] [DOI] </nowiki>
- ↑ <pubmed>16935554</pubmed>
- ↑ 16.0 16.1 </nowiki>
- ↑ <pubmed>15020481</pubmed>
- ↑ <pubmed>15831788</pubmed>
- ↑ <pubmed>17578919</pubmed>
- ↑ <pubmed>11707339</pubmed>
- ↑ <pubmed>12095618</pubmed>
- ↑ <pubmed>21283790</pubmed>
- ↑ <pubmed>10537186</pubmed>
- ↑ <pubmed>25121765</pubmed>
- ↑ <pubmed>21701686</pubmed>
- ↑ <pubmed>X</pubmed>
- ↑ 27.0 27.1 </nowiki>
- ↑ <pubmed>10320586</pubmed>
- ↑ <pubmed>18215304</pubmed>