General Information/IS Organization
Contents
General
In addition to being small, insertion sequences are genetically compact (Fig.18.1). They generally encode no functions other than those involved in their mobility although individual members of several families which include additional genes are now being identified. IS-encoded functions include factors required in cis, in particular recombinationally active DNA sequences that define the ends of the element together with an enzyme, the transposase (Tpase), which recognises and processes these ends. The Tpase is generally encoded by a single or perhaps two, open reading frames and consumes nearly the entire length of the element.
Terminal inverted repeats
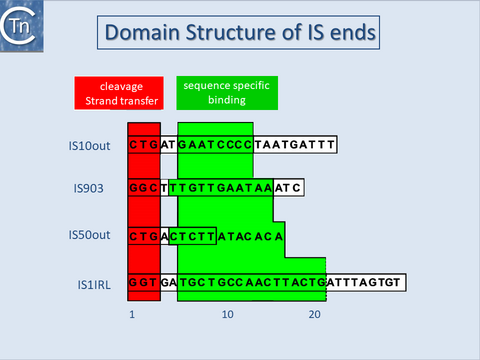
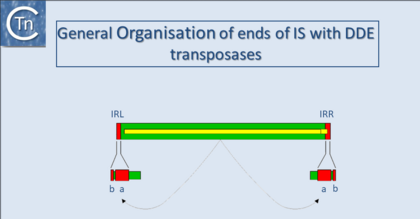
With several notable exceptions (the IS91, IS110 and IS200/605 families; TABLE Characteristics of IS families) the majority of ISs exhibit short terminal IR of between 10 and 40 bp. In those cases examined experimentally, the IRs can be divided into two functional domains (Fig.11.1). Domain “b” includes the two or three terminal base pairs (Fig.18.2), and is involved in the cleavages and strand transfer reactions leading to transposition of the element. Domain “a” is positioned within the IR and is involved in Tpase binding[1][2][3][4][5][6][7].
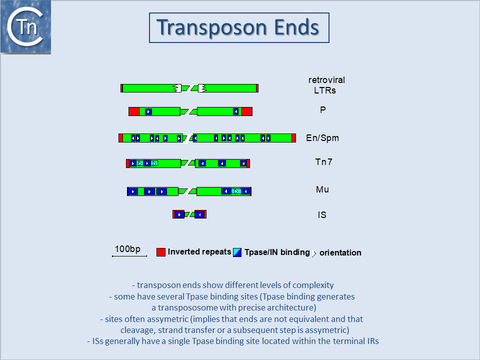
A similar organization has also been proposed for the transposon Tn3[8] and for the related γδ transposon[9]. The simple single terminal Tpase binding sites of ISs are to be contrasted with the multiple and asymmetric protein binding sites observed in the case of bacteriophage Mu[10] and transposons Tn7[11], and probably Tn552[12][13]. Multiple protein binding sites are also a characteristic of the complex En/Spm and Ac elements of maize (see [14][15] (Fig.18.3). It is worth noting that members of the IS21 family also carry multiple repeated sequences at both ends which may also represent Tpase binding sites[16][17].
By accommodating different binding patterns at each end, such an arrangement can provide a functional distinction between the ends either in the assembly or in the activity of the synaptic complex. In addition, indigenous IS promoters are often located partially within the IR sequence upstream of the Tpase gene, by convention IRL. This arrangement may provide a mechanism for autoregulation of Tpase synthesis by Tpase binding. Binding sites for host specified proteins are also often found within or close to the terminal IRs and these proteins may play a role in modulating transposition activity or Tpase expression.
Domain structure of transposases
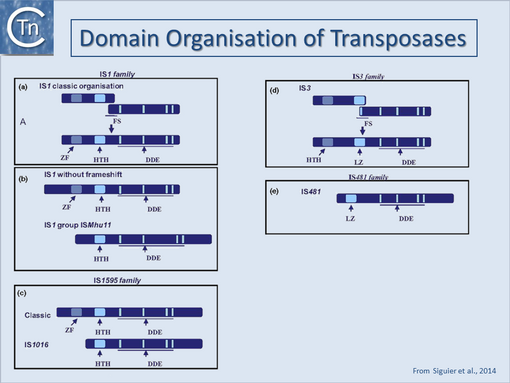
A general pattern for the functional organisation of Tpases appears to be emerging from the increasing number which have been analysed. Many can be divided into topologically distinct structural domains and, although several regions of the protein may contribute to a given function, the isolated domains themselves often exhibit a distinct function. The sequence-specific DNA binding activities of the proteins are generally located in the N-terminal region while the catalytic domain is often localised towards the C-terminal end: IS1[18][19]; IS30[20];Mu, (see [21][22]; Tn3[23][24]; IS50[25]; IS903[26]; IS911[27]; for a review see [28] (Fig.18.4). One functional interpretation of this arrangement for prokaryotic elements is that it may permit interaction of a nascent protein molecule with its target sequences on the IS thus coupling expression and activity. This notion is reinforced by the observation that the presence of the C-terminal region of the IS50, IS10 and IS911 Tpases appears to mask the DNA binding domain and reduce binding activity[29][30] possibly by masking the DNA binding domain. This arrangement might favor activity of the protein in cis, a property shared by several Tpases (see Activity in cis). Similar masking appears to occur with the IS1 (D. Zerbib and M. Chandler, unpublished) and the IS911[31][7] Tpases. In several cases these domains are assembled into a single protein from consecutive orfs by translational frameshifting (Programmed translational frameshifting). In the case of IS911, it has been demonstrated that transposase binding to the IS ends occurs as the protein is translated[32].
One exception to this is the transposase of the IS110 family which encodes a DEDD transposase closely related to the RuvC Holiday resolvase (see [33][34]) and in which the catalytic domain appears to precede the DNA binding domain.
In addition to functional domains for DNA binding and catalysis, many, if not all transposases have the capacity to generate multimeric forms essential for their activity (see [28][35]). This is true of prokaryotic elements such as bacteriophage Mu (see [36]), IS50[37], IS911[31][7][35], IS608 and ISDra2[38][39] (but apparently not IS10[40], and of eukaryotic elements such as the retroviruses (see [41][42][43][44]) whose integrase (IN) (transposase) appears to be a dimer of dimers both with and without DNA bound[43][45][46][47] as does the purified P element transposase[48], the mariner-like element, Mos1[49][50][51] and hermes (which appears to be an octomer)[52][53][54]. With the results of an increasing number of structural studies of these types of enzyme, it will be of great interest to compare the overall similarities of equivalent functional domains as has been recently possible with the catalytic domains of retroviral integrases, Mu transposase and other polynucleotidyl transferases such as the Holiday resolvase, RuvC and RnaseH (see [55][56]). One particular type of structure, a leucine zipper, clearly plays a vital role in multimerization of a number of transposases[31][35][57][58][59][60][61][62].
Finally, several DDE transposases exhibit what may be thought of as an “orphan” domain located between the second D and final E of the DDE motif[63] (Table transposases examined by secondary structure prediction programs). These can be either largely α-helical or β-stranded. It is likely that these insertion domains play subtle roles in the chemical pathways involved in the transposition of their cognate transposable elements. The largely β-stranded IS50 (Tn5) associated insertion domain (Major DDE transposition pathways - Fig.7.3)) interacts with and assists in stabilizing a hairpin-loop structure at the transposon end formed as an intermediate during transposition[64][65][66] and therefore performs a crucial function. Another type of insertion domain is present at the same topological position in the eukaryotic Hermes transposase[52]. This long insertion domain is entirely α-helical. In contrast to Tn5 whose hairpin intermediate is formed on the transposon end, Hermes transposes using an intermediate in which a hairpin is formed on the flanking donor DNA[67][68]. It appears that the insertion domain assists stabilization of the Hermes hairpin intermediate. A similar α-helical insertion has been identified in the VDJ recombinase, RAG1[67] which also generates a hairpin intermediate similar to that of Hermes. In Hermes, the domain also assists in forming transposase multimers.
Direct target repeats
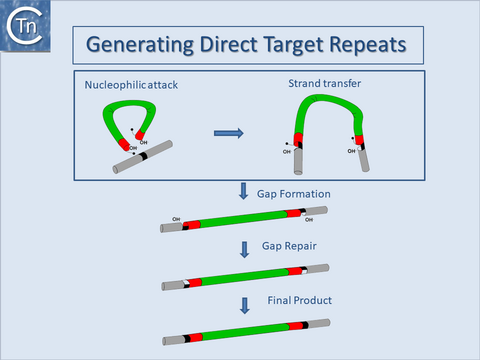
Another general feature of IS elements is that, on insertion, most generate short directly repeated sequences (DR) of the target DNA flanking the IS. The attack of each DNA strand at the target site by one of the two transposon ends in a staggered way during insertion provides an explanation for this observation. The DR is generated by repair of the “gap” between cleavage sites (Fig.18.5).
The length of the DR, generally between 2 and 14 bp, is characteristic for a given element and a given element will generally generate a duplication of fixed length. This is determined by the architecture of the transposition complex or transpososome which imposes constraints on the distance between cleavages on each strand of the target DNA[69][63][70].
However, certain ISs have been shown to generate DRs of atypical length at a low frequency, presumably reflecting small variations in the geometry of the transpososome (see [71]). Although some notable exceptions exist in which there is a systematic absence of DRs (either within a given family or in several independent transposition events of a given element), care should be taken in interpreting the absence of DRs in isolated cases.
A lack of DRs can simply result from homologous inter- or intra-molecular recombination between two IS elements, each with a different DR sequence. This would result in a hybrid element carrying one DR of each parent. It can also arise from the formation of adjacent deletions resulting from duplicative intramolecular transposition. In this case, a single copy of the DR is located on each of the reciprocal deletion products (see for example [72][73] or, more recently in a clinical context, [74].
Three IS, IS1549, IS1634 and IS1630, have been identified which appear to generate long DRs of quite variable length[75][76][77]. Two, IS1549 and IS1634, are distantly related to the IS4 family and one, IS1630, belongs to the IS30 family. The mechanism involved in generating such long DRs is at present unknown. However, it seems reasonable to propose that the target DNA may be able to form a loop within the transpososome and thereby bringing somewhat distant phophodiester bonds into close proximity.
Bibliography
- ↑ <pubmed>8898394</pubmed>
- ↑ <pubmed>2161528</pubmed>
- ↑ <pubmed>2551675</pubmed>
- ↑ <pubmed>6306482</pubmed>
- ↑ <pubmed>2832849</pubmed>
- ↑ <pubmed>28776821</pubmed>
- ↑ 7.0 7.1 7.2 Normand C, Duval-Valentin G, Haren L, Chandler M . The terminal inverted repeats of IS911: requirements for synaptic complex assembly and activity. - J Mol Biol: 2001 May 18, 308(5);853-71 [PubMed:11352577] [DOI] </nowiki>
- ↑ <pubmed>2155858</pubmed>
- ↑ <pubmed>7723015</pubmed>
- ↑ <pubmed>6094016</pubmed>
- ↑ <pubmed>8556868</pubmed>
- ↑ <pubmed>2170815</pubmed>
- ↑ <pubmed>7828593</pubmed>
- ↑ <pubmed>8556865</pubmed>
- ↑ <pubmed>8556866</pubmed>
- ↑ <pubmed>11125167</pubmed>
- ↑ <pubmed>9729608</pubmed>
- ↑ <pubmed>2553980</pubmed>
- ↑ <pubmed>2162466</pubmed>
- ↑ <pubmed>2154486</pubmed>
- ↑ <pubmed>8556870</pubmed>
- ↑ <pubmed>8646783</pubmed>
- ↑ <pubmed>8382339</pubmed>
- ↑ <pubmed>8080658</pubmed>
- ↑ <pubmed>8289277</pubmed>
- ↑ <pubmed>9417930</pubmed>
- ↑ <pubmed>8950268</pubmed>
- ↑ 28.0 28.1 Haren L, Ton-Hoang B, Chandler M . Integrating DNA: transposases and retroviral integrases. - Annu Rev Microbiol: 1999, 53;245-81 [PubMed:10547692] [DOI] </nowiki>
- ↑ <pubmed>8057357</pubmed>
- ↑ <pubmed>8412678</pubmed>
- ↑ 31.0 31.1 31.2 Haren L, Polard P, Ton-Hoang B, Chandler M . Multiple oligomerisation domains in the IS911 transposase: a leucine zipper motif is essential for activity. - J Mol Biol: 1998, 283(1);29-41 [PubMed:9761671] [DOI] </nowiki>
- ↑ <pubmed>22195971</pubmed>
- ↑ <pubmed>15866929</pubmed>
- ↑ <pubmed>12897009</pubmed>
- ↑ 35.0 35.1 35.2 </nowiki>
- ↑ <pubmed>8805293</pubmed>
- ↑ <pubmed>7958902</pubmed>
- ↑ <pubmed>16209952</pubmed>
- ↑ <pubmed>20890269</pubmed>
- ↑ <pubmed>8565068</pubmed>
- ↑ <pubmed>7526778</pubmed>
- ↑ <pubmed>1322888</pubmed>
- ↑ 43.0 43.1 Bao KK, Wang H, Miller JK, Erie DA, Skalka AM, Wong I . Functional oligomeric state of avian sarcoma virus integrase. - J Biol Chem: 2003 Jan 10, 278(2);1323-7 [PubMed:12446721] [DOI] </nowiki>
- ↑ <pubmed>15718297</pubmed>
- ↑ <pubmed>17157316</pubmed>
- ↑ <pubmed>19609359</pubmed>
- ↑ <pubmed>19229293</pubmed>
- ↑ <pubmed>17644523</pubmed>
- ↑ <pubmed>8913752</pubmed>
- ↑ <pubmed>17565190</pubmed>
- ↑ <pubmed>19766564</pubmed>
- ↑ 52.0 52.1 </nowiki>
- ↑ <pubmed>16511103</pubmed>
- ↑ <pubmed>25036632</pubmed>
- ↑ <pubmed>8696976</pubmed>
- ↑ <pubmed>8548793</pubmed>
- ↑ <pubmed>2147779</pubmed>
- ↑ <pubmed>8647395</pubmed>
- ↑ <pubmed>22032517</pubmed>
- ↑ <pubmed>8643520</pubmed>
- ↑ <pubmed>19416360</pubmed>
- ↑ <pubmed>8520113</pubmed>
- ↑ 63.0 63.1 Dyda F, Chandler M, Hickman AB . The emerging diversity of transpososome architectures. - Q Rev Biophys: 2012 Nov, 45(4);493-521 [PubMed:23217365] [DOI] </nowiki>
- ↑ <pubmed>10884228</pubmed>
- ↑ <pubmed>15102449</pubmed>
- ↑ <pubmed>17176076</pubmed>
- ↑ 67.0 67.1 </nowiki>
- ↑ <pubmed>30239795</pubmed>
- ↑ <pubmed>16181782</pubmed>
- ↑ <pubmed>21439812</pubmed>
- ↑ <pubmed>26104718</pubmed>
- ↑ <pubmed>6314502</pubmed>
- ↑ <pubmed>7489730</pubmed>
- ↑ <pubmed>26060276</pubmed>
- ↑ <pubmed>10601219</pubmed>
- ↑ <pubmed>9495740</pubmed>
- ↑ <pubmed>9973360</pubmed>